Rhizosphere
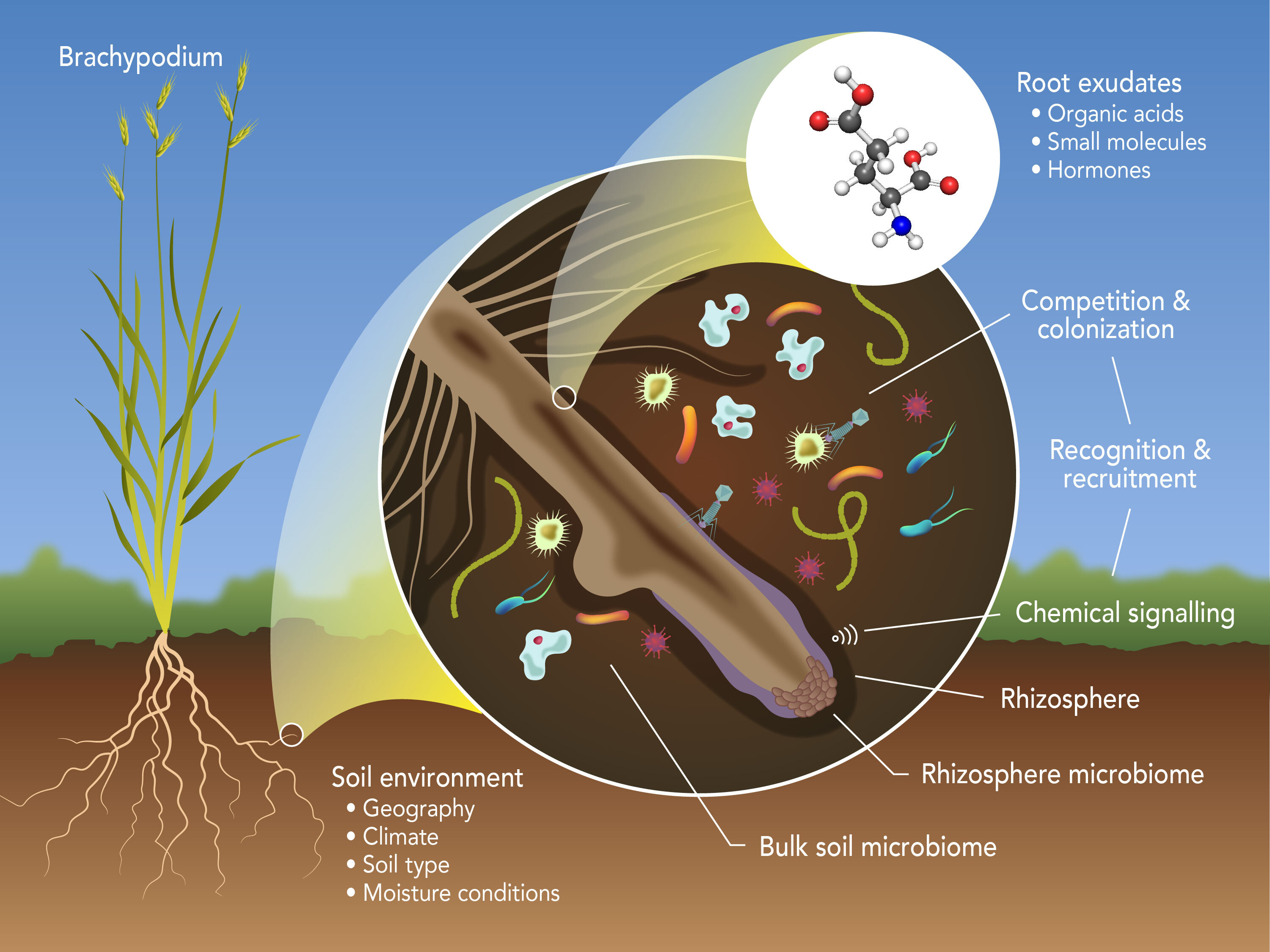
The rhizosphere is the interface between the root system of a plant and its surrounding soil. The microbiome of the rhizosphere, which is the totality of all microbes present there, represents a complex microbial ecosystem that nourishes the terrestrial biosphere. The rhizosphere is the interface between the root system of a plant and its surrounding soil. It is arguably the most complex terrestrial microbial habitat on Earth, supporting an estimated 1011 microbial cells per gram root , with 1012 functional genes per gram soil. The plant-specific microbial community of the rhizosphere, collectively referred to as the rhizosphere microbiome, is fueled by plant root exudates and contains a vast diversity of microorganisms (e.g., archaea, bacteria, eukaryotes, and viruses). The composition and relative abundance of the plant root exudates have a profound impact on the surrounding soil environment and, hence on the rhizosphere microbiome. Reciprocally, the structural and functional properties of the rhizosphere microbiome affect plant growth and fitness. To untangle the complexity of the rhizosphere, and of the rhizospheric microbiome in particular, an integrated multiomics approach can be applied to reveal the composition of the rhizospheric microbiome (through 16S ribosomal amplicons and metagenomics), the functional properties of the microbiome (through metatranscriptomics and metaproteomics), and the signaling network within the rhizosphere (through metametabolomics).
Rhizosphere - Carbon
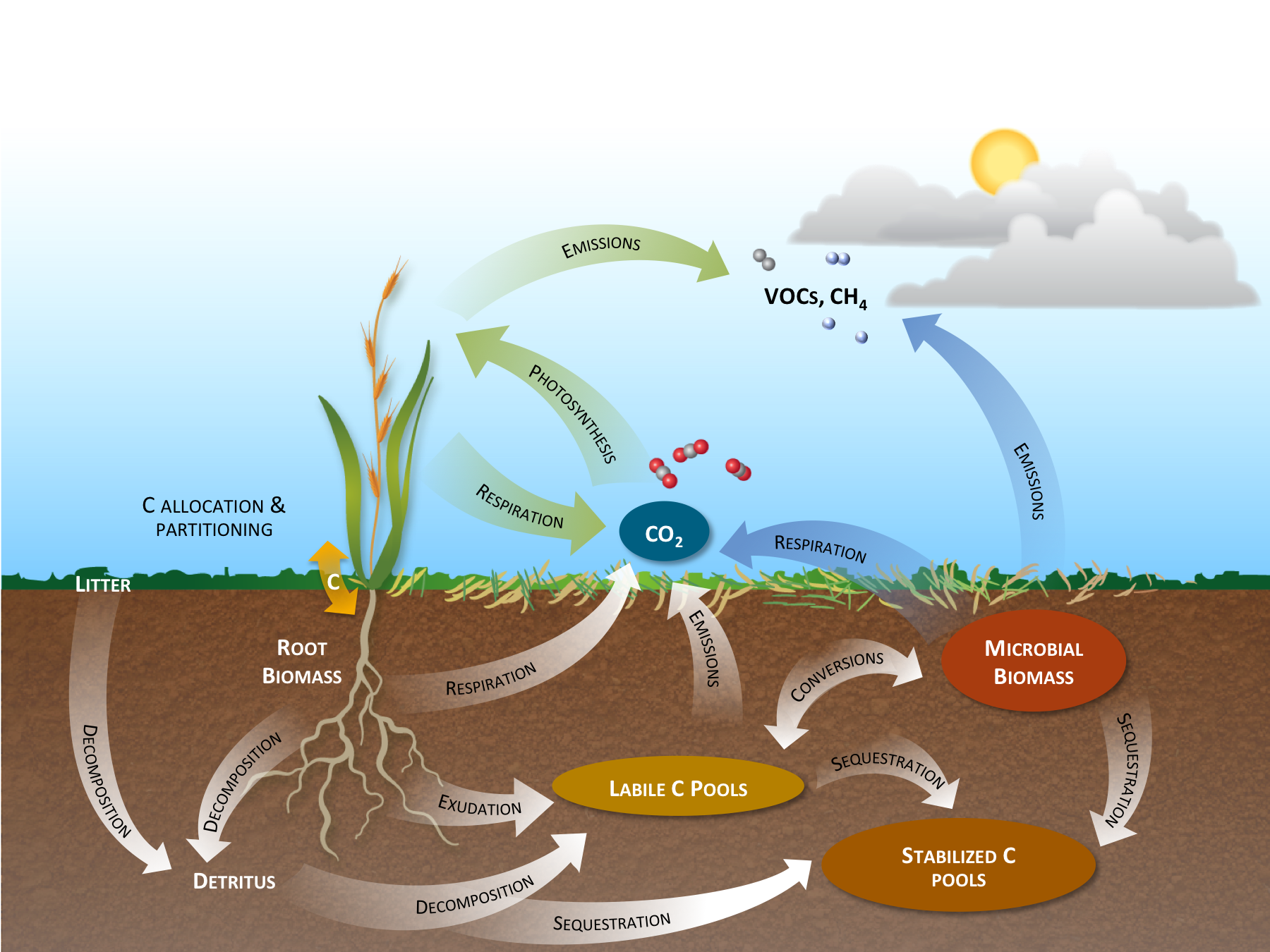
Interactions between the rhizosphere and other components of the plant ecosystem. Carbon (C) enters the soil as root exudates or via decomposition of root or aboveground biomass. In the soil, C exists in root or microbial biomass, as bioavailable labile organic C, or as more recalcitrant C. Carbon exits the soil as direct emissions, or via root or microbial respiration, with microbial-mediated soil respiration being the major source of CO2 from terrestrial ecosystems. Carbon is also lost from the ecosystem as volatile organic compounds (VOCs) and methane (CH4).
The rhizosphere as a surrogate model for the human microbiome.
Clinical, animal, and patient-related work reveal the human host interacting with its gut microbiome, but this is not the whole story of microbiomes. To get that we can turn to the rhizosphere, the interface between plant roots and the surrounding soil. The rhizosphere contains all domains of life, and represent the most diverse and dynamic microbiomes on the planet (White III et al., 2017, Rhizosphere). Humans and global ecosystems are dependent on nutrients and nutrition provided by this millimeter-scale interface between plant microbes and soil. While the rhizosphere has been studied for over a hundred years, little is known about the trans-kingdom interactions between plant-microbes and viruses, for instance. The rhizosphere represents the “gut” of a plant and is a surrogate model for the human gut microbiome. We developed omics technologies and software to study this complex and methodologically challenging ecosystem.
Why so challenging? First, soil/rhizosphere metagenomics have been plagued by poor functional and taxonomic annotation because of very short-read sequencing technologies. To remedy this problem, we applied Moleculo long-read sequencing technology, in which Dr. White III was involved in the initial development at Stanford University (Voskoboynik, White III, et al., 2013, ELife) and when sequencing the Botryllus schlosseri genome via soil metagenomics of Kansas prairie soil (White III et al., 2016, Msystems). Moleculo sequencing has provided the most contiguous assembly of soil metagenome (i.e., >3,000 contigs >10 kbp), and the first proof-of-concept of bacterial genomes directly assembled from a soil metagenome. The Moleculo assembly provided a robust database for searching metaproteomic data from complex soils, which in turn allowed for further technology development regarding protein extraction from soil (Nicora, White III et al., 2018, JOVE) and better fractionation of proteins for the highest possible coverage of the metaproteome of soil (Callister, White III et al., 2018, Soil Biol Biochem).
The Moleculo sequencing required new software for assembly, annotation, metagenomic genome binning, and coverage analysis which led to the creation of ATLAS, a comprehensive tool for metagenomic/metatranscriptomic analysis (White III et al., 2017, PeerJ). Mercat has provided a homology/alignment-free method to direct comparisons for large complex metagenomes from the soil—all in a tool that runs on a standard laptop (White III et al., 2017, PeerJ). ATLAS was used disentangle the complexity of permafrost soil in the high Arctic in a recent collaboration (Müller, White III et al., 2018, Environ Microbiol).
As with the nexus of the human gut-oral microbiome, even less is known about phage and viruses within soil and rhizosphere ecosystems. In marine ecosystems, viruses have been estimated to lyse around 20% of the biomass daily. This viral lysis results in the 'viral shunt' of marine food webs, which diverts carbon and nutrients from secondary consumers and creates a vast dissolved organic carbon (DOC) pool in the ocean. Phage in marine systems have been called 'puppet masters' for their role in the fate of marine bacteria. Cyanophage (that is, the phage of cyanobacteria) have accessory metabolic genes (AMGs) that directly manipulate host carbon cycling by blocking Calvin cycle to preferentially redirect host metabolism towards nucleic synthesis, and by increasing energy metabolism via photosynthesis using viral psbA genes.
It has been discussed that a warming climate will make soil “breathe harder,” releasing more carbon dioxide into the environment as for instance frozen carbon cools are thawed and microbial heterotrophy increases. Meanwhile, the role of viruses in terrestrial ecosystems is a vast unknown.